Environmental Impacts on the Epigenome
 DMR overlaps
DMR overlaps
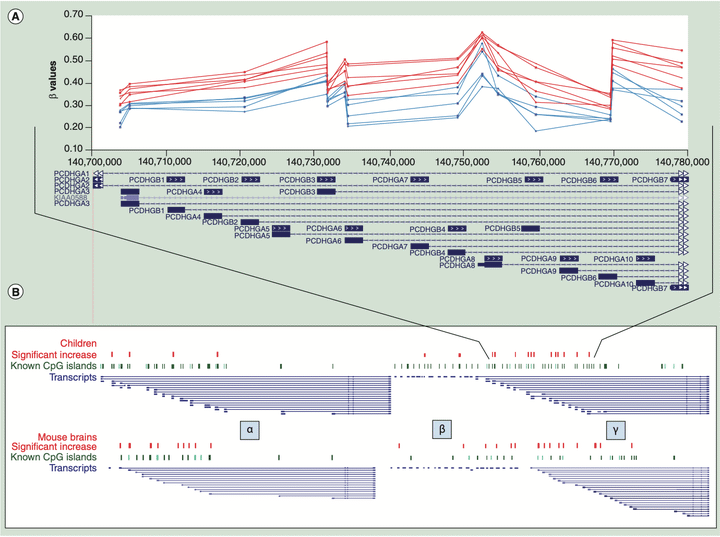
The consumption of alcohol during any stage of pregnancy can result in fetal alcohol spectrum disorders (FASDs), with there being no empirically determined safe dose or safe timing of prenatal alcohol exposure (PAE). FASDs are the leading preventable cause of intellectual disability in the western world. In North America, ~10% of pregnant women and ~20% of breast-feeding women consume alcohol; however, FASDs are diagnosed in only ~2-3% of children. Accurate biomarkers of PAE and its neurodevelopmental impacts are needed, as current diagnostics are not specific to the large number of individuals on the spectrum without craniofacial malformations who require specialized behavioral therapies specific to FASDs. Since some FASD phenotypes can be attenuated by the supplementation of methyl donors from the one-carbon metabolism pathway, DNA methylation plays a key role in the etiology of FASDs. Furthermore, FASDs represent one of a select few common NDDs where the cause is known (PAE), which poises it to provide generalizable insights for NDDs with an environmental component. During my graduate research, we examined a mouse model of a chronic but moderate prenatal alcohol exposure throughout pregnancy. I utilized microarray technologies to integrate genome-wide DNA methylation, microRNA expression, and gene expression profiles of exposed mouse brains at adulthood. Strikingly, while there were genome-wide alterations at all levels, there was a distinct profile of alterations to genomically imprinted clusters of non-coding RNA, which are expressed in a parent-of-origin specific manner and are involved in (neuro)development. These findings were then replicated in other mouse models of PAE, both in collaboration with other members from my graduate lab, as well as by independent laboratories. After establishing that there are long-term alterations to the epigenome in the brains of prenatal alcohol exposed mice, we formed a collaboration with Dr. Joachim Kapalanga (MD-PhD). I examined the DNA methylation profile of cells in the buccal swabs of young children with FASD using the 450K BeadChip array and compared the results to our mouse model. Interestingly, we observed similar alterations to epigenetically sensitive regions that were identified in the rodent models. This work was the first epigenome-wide investigation of children with FASD and has provided the foundation for the possibility of a molecular diagnostic for FASD. The results implicated differentially methylated regions (DMRs) within the clustered protocadherins, which establish single-cell neuronal identity, and this result was seen in both adult mouse brains and the buccal swabs of children. Notably, buccal epithelial cells are derived from the neuroectoderm and thus share a close ontogenetic relationship with brain, which may explain the shared epigenomic profile of early neurodevelopment. An independent laboratory has replicated the large-scale protocadherin DMRs and noted the effect of genetic variation in a cohort of children from different ethnicities.